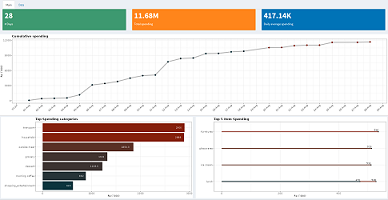
Dashboard: Expense tracking
Shiny dashboard to track finances with an input file
Data professional since 2014 with experience across manufacturing, education, academic, healthcare technology, and insurance. Platform-agnostic, with experience working with AWS and Microsoft Azure environments. Skilled in programming and statistical languages, regularly writing scripts and queries and deploying models in R, SQL, Bash and Python.
My technical experience spans the roles of a data engineer, data analytics, and a data scientist. Professionally I have experience implementing data governance framework in organisations spanning the scale of a start-up to corporates.
I am also a Google-certified data analytics professional.

Shiny dashboard to track finances with an input file
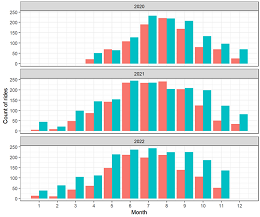
End-to-end analytics workflow from data extraction to business insights and recommendations based on data from the bike-sharing industry provided by Motivate International Inc.

Tutorial on how to automatically move each file into their designated subdirectories in R.
Cover image taken from Domiriel
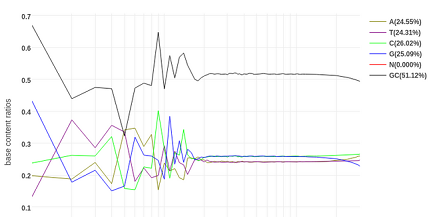
The first of a two-part tutorial on biomarker screening and pan-genome comparison of bacterial whole genome sequences as held at UM 30-1 December 2022. Covers working your way around Ubuntu and the Bash programming language.
Skills: Linux Bash, Ubuntu, genomics, bioinformatics
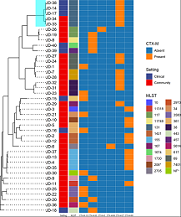
The second of a two-part tutorial on biomarker screening and pan-genome comparison of bacterial whole genome sequences as held at UM 30-1 December 2022. Covers the use of R statistical language for data wrangling, transformation, and visualisation.
Skills: R, data wrangling, data transformation, data visualisation

Insert imageJ python script for looping image processing here
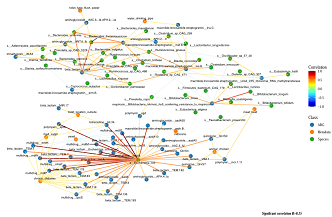
Published peer-reviewed article correlating E. coli and various lifestyle factors with antibiotic-resistant bacteria colonisation
Skills: Linux Bash, Ubuntu, risk factor analysis, genomics, bioinformatics, statistics, data visualisation
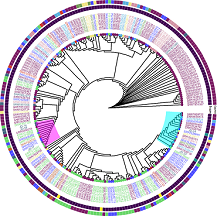
Published peer-reviewed article profiling antibiotic-resistant E. coli in a Malaysian community through whole-genome sequencing
Skills: Linux Bash, Ubuntu, genomics, bioinformatics, statistics, data visualisation, phylogenetic comparison
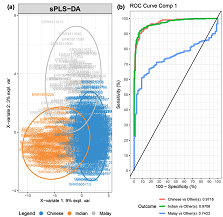
Published peer-reviewed articles on the development and validation of a random forest model capable of predicting ethnic origin based on the human gut microbiome.
Skills: Machine learning, correlation, ordination, meta-analysis, data mining, bioinformatics, metagenomics
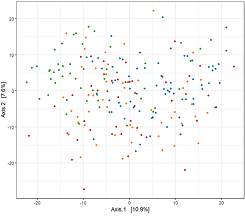
Published peer-reviewed articles profiling the gut microbiome profile and predicted functional pathways across across ethnicity
Skills: Machine learning, ordination, differential abundance analysis, statistics, bioinformatics, metagenomics
Interested in my skills and what I can offer? Get in touch!